Implementation: overview
These pages document the implementation of the numerical experiments for paper “On Aligning Non-Order-Associated Binary Decision Diagrams” by Bochkarev and Smith. The notes along with the accompanying source code and data are assumed to provide enough information to:
inspect the raw data,
reproduce / adjust / inspect the figures from the paper (given the experiment log files) as necessary, and
completely reproduce the paper results from scratch, including instance generation.
It is our special interest to help the reader build upon our results, so please do not hesitate to address any technical questions to Alexey Bochkarev.
Computational infrastructure
To run the experiments we used machines running GNU/Linux system (remote computational resources provided by Clemson Palmetto cluster and laptops), running the following software:
experiments implementation: python3.
packages list, the usual way should work:pip install -r requirements.txt
figures: R. To install (a superset of) the necessary libraries, from within R session:
install.packages(c("dplyr", "ggplot2", "ggridges",
"gridExtra", "latex2exp", "optparse", "reshape",
"stringr", "tidyr", "viridis", "xtable"))
parallel computation: GNU parallel,
experiments pipeline: GNU make and PBS (for job scheduling on the cluster).
(we also used a separate tool to receive the results promptly via Telegram, but this can be safely removed)
numerical experiments require Gurobi solver.
most graphs were visualized used a stand-alone program called Graphviz Note that before
pip install pygraphvizyou might need to install the program and the header files as well. On my Debian distribution I needed to dosudo apt install graphviz graphviz-dev.
Note
For the (python) environment management we used the recommended venv-way. For example, the virtual environment can be created and activated with:
$ python3.10 -m venv .venv $ source .venv/bin/activate $ # then, we can install the necessary packages there with: $ pip install -r requirements.txt
An equivalent for conda-based system would be:
$ conda create --name py310 python=3.10 $ conda install --name py310 --channel conda-forge --file requirements.txt $ conda activate py310
(However, we did not use conda. If you do – please, let us know if it works as well.)
General reproducibility
Assuming the necessary software is present, one can rebuild any figure from the
existing log file (specific experiment results) by running a make command
from the root project directory (align-BDD). For example, this
must work (assuming the necessary dependencies are installed):
make figures/simpl_heuristics.eps # to rebuild Figure 6, or
make figures # to rebuild all figures for the paper
GNU make will consider files’ timestamps and re-run the experiments as necessary (e.g., if the instance generation procedure has changed). To force a complete run (generate all the data, make all experiments, and build all figures), just clean up the previous results first:
make fresh # to delete all instances and run logs
make figures # to rebuild everything
Note, however, that depending on the parameters (which are set in Makefile)
and the hardware, it might take quite some time to recalculate everything. PBS
scripts that were used to run our experiments on the computation cluster are
kept in folder 📁 pbs. Certainly, they are dependent on the hardware and the
available software, but these materials hopefully will provide a good starting
point to reproduce the results presented in the paper using a PBS-based system.
All the recipies (how to make each figure or other artifact) are described in
the file called Makefile, which is basically a list of commands and source
files needed to build each target.
Project repository structure.
All the source data is available in the project repository.
Key folders are:
📁 instances: all the instance (“raw”) data in a compressed format (tar.gz; see e.g. here if unsure how to open it),📁 run_logs: all the numerical results – “run logs,” mostly as comma separated files (.csv)📁 experimentsand the root folder contain all key implementation files (in Python).📁 post_processing: the R code that produces figures from the run logs.📁 figures: the key figures themselves.📁 docs: the code documentation source (i.e., these pages).📁 tests: most of the code testing (for python code).📁 pbs: auxiliary PBS scripts used to run the code on the computational cluster.
The computations are summarized in the following table (scrollable right):
Figure / Table |
Filename |
Experiments code (scripts) |
Raw instances |
Format |
Experiment run log |
Plotting code |
|---|---|---|---|---|---|---|
Table 1 |
|
|
BDD |
|
|
|
Figure 6 |
|
|
||||
Figure 7 |
|
|
||||
Figure 17 |
|
|
|
|||
Figure 19 |
|
|
|
|
||
Figure 8 |
|
|
|
|
||
Figure 20 Figure 21 Figure 22 Figure 23 |
|
|
|
|
||
Figure 9b Figure 10a Figure 10b |
|
|
json |
|
|
|
Figure 11 |
|
|
json |
|
|
Figure 17: sample branch and bound tree was generated by experiments.sample_BB_tree.
Figures 1-5, 9a, 12-16, and 24-27 do not involve any computations.
Key ‘global’ parameters (instance sizes, generation parameters, etc.) are set in Makefile. (The rest are specified in the respective python files in experiments folder.)
Implementation details
Here are implementations of the key algorithms from the paper:
Reference |
Brief Description |
Implementation |
|---|---|---|
Sec. 2.1 |
BDD data structure implementation |
|
Sec. 3.2 |
Weighted variable sequence ( |
|
Algo 3 |
A |
|
Algo 4 |
Align-to (weighted variable sequence, linear time)
(Algo 1 is slower and is not used in the experiments)
|
|
Algo 5 |
Branch-and-bound algorithm for the simplified problem.
- lower bound:
- upper bound:
|
|
Sec. 4.1.2 |
Various heuristics for aligning BDDs and VarSeqs. |
|
Algo 6 |
Creating a BDD encoding an UFLP sub-instance.
(for a given order of variables)
|
|
Example 5
(App. H)
|
Finding a good order of variables
for an UFLP sub-instance.
|
|
Sec. 4.2 |
Solving j-UFLP:
- with naive MIP (formulation (2)):
- CPP as an MIP:
- CPP / shortest-path over intersection BDD:
|
See
UFLP_2_cav for the overview, and then, in particular: |
Raw data formats
Binary Decision Diagrams
- A
.bddfile represents a structured plain-text description of a BDD explained in more detail in
BDD.BDD.save(). Any diagram (say, from filefilename.bdd) can be loaded for inspection with the code like:
import BDD
A = BDD.BDD()
A.load("./filename.bdd")
A.show() # will pop up the diagram if the relevant software is installed
Note that each align-BDD instance is just a pair of BDDs with the same number.
For example, instance number one from orig_problem.tar.gz (which ends up,
for example, in Figure 7) is contained in two files, A1.bdd and B1.bdd.
j-UFLP instance data
A .json file describes a j-UFLP instance in JSON format.
For more details on how exactly it is saved, see jUFLP_utils.save_inst().
Each instance can be loaded for inspection from a file as follows, using the
auxiliary code we have in jUFLP_utils:
import json
from jUFLP_utils import load_inst, draw_jUFLP_inst
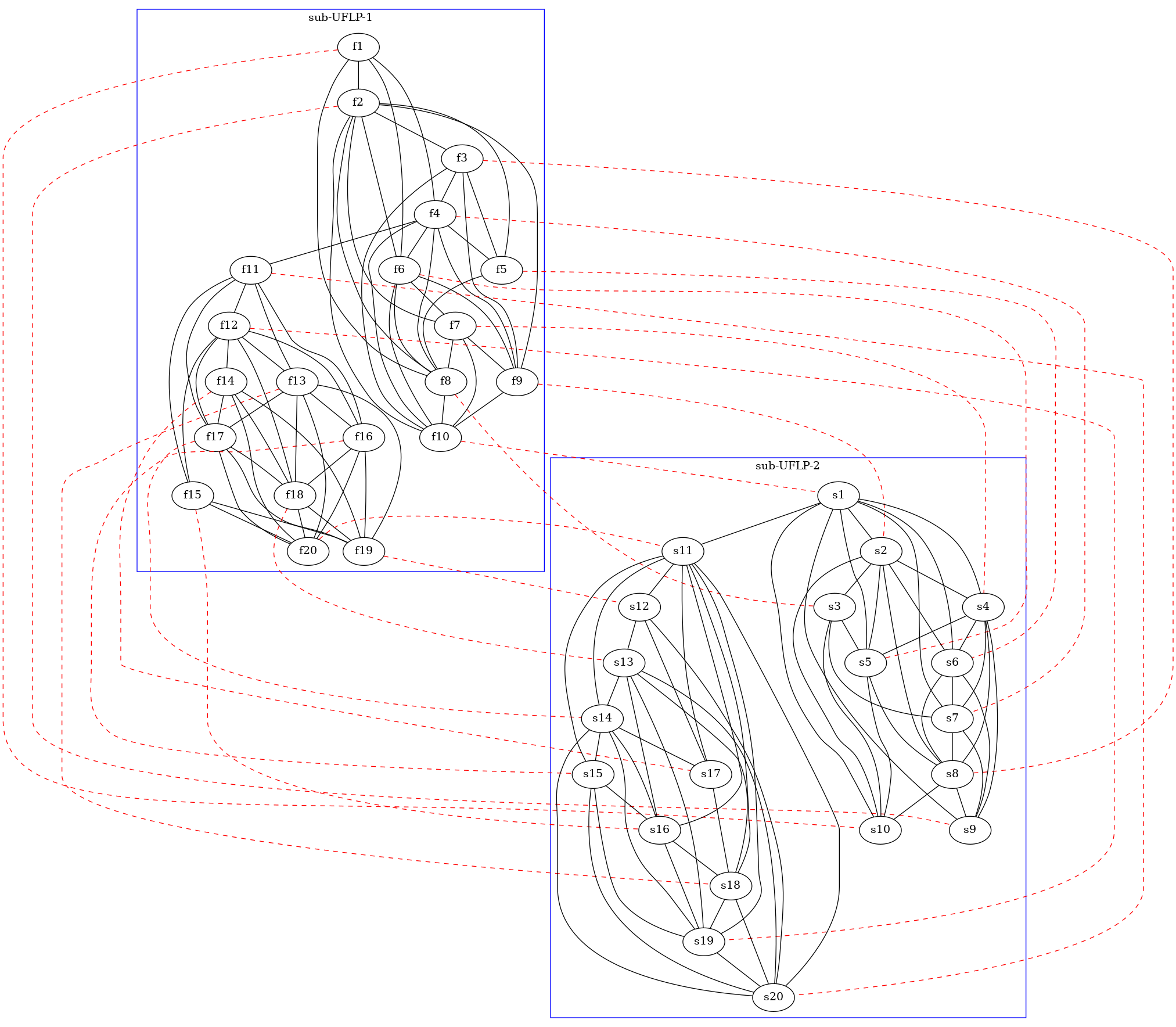
i1, i2, link = load_inst("./instances/jUFLP_cm/inst_1.json")
draw_jUFLP_inst(i1, i2, link, filename="tmp/jUFLP-check.dot")
After this, variables i1, i2, and link will contain the j-UFLP
instance description (respectively, the two sub-instances and the linking
dictionary). Moreover, function jUFLP_utils.draw_jUFLP_inst() will
create a .dot (Graphviz) file visualizing the instance as a graph,
similar to Figure 9a. So that invoking dot -Tx11 tmp/jUFLP-check.dot
(on my GNU/Linux machine) gives something like:
Code organization
The code is organized into several blocks (modules) as follows.
Producing figures
The code that physically produces figures from CSV run logs is written in R (and the wonderful tidyverse/ggplot2 library). The code is conceptually simple and,
hopefully, self-explanatory – see .R - files in 📁 post_processing.
Key data structures and algorithms.
The core data structures critical to the paper are presented in the following modules (click on the links for more implementation details and the source code):
Implements the Binary Decision Diagram data type. |
|
Implements weighted variable sequences data structure. |
|
Implements several heuristics to be used with a separate script to generate |
|
A branch-and-bound search scheme implementation for the align-sequences problem: alignts two |
The code relevant to j-UFLP application (Section 4.2) is here:
Implements basic functions for UFL -- Uncapacitated Facility Location. |
|
Solves UFLP using a DD-based approach. |
|
Implements an algorithm to find a var order for BDD representing an UFLP. |
|
Proof-of-concept experiment: joint UFLP over special class of instances. |
|
Utility / helper functions for j-UFLP: used save / load / draw instances. |
Numerical experiments
Description for specific problems serves the basis for further numerical
experiments, which are implemented as separate python programs in 📁
experiments
Generates a pair of random BDDs ("original problem" instance). |
|
Solves given align-BDD instances to benchmark different methods. |
|
Implements most of the numerical experiments in the paper. |
|
Generates and solves the special class j-UFLP instances. |
|
Contains the j-UFLP instance generation code (along with some legacy experiments). |
Testing framework
Key implementations are covered with tests, mostly using pytest framework. On top of the testing code in the files
mentioned above (testing functions have test_ prefix in the name), some
tests are in 📁 tests:
Tests some key procedures for |
|
Tests equivalence of BDDs after operations. |
|
Tests the |
|
Tests BB search correctness ( |
|
Tests the UFLP-related machinery ( |